GMM trajectory statistics and figures for quadratic models
#source raw data directory: data_raw and data included
source("../../../isolation_trajectories_data_path.R")Quadratic GMM
Quadratic term significance for each model
si.quad.all <- readModels(paste0(mplus_GMM_clustered_full_output_QUAD_data_path), recursive = TRUE)#extract the summary variables from the mplus output files. Assuming there are multiple files in the above directory, model summaries could be retained as a data.frame as follows:
si_quad_summaries <- do.call("rbind.fill",
sapply(si.quad.all,
"[",
"summaries"))Here we only reprt trsults for the two and three class models as the four to six class models did not converge.
- 4th class originally had problems with negative S variances - set S@0 for this model, this then resulted in non-convergence.
- 5th class originally had problems with negative S variances - set S@0 for this model, this then resulted in non-convergence.
- 6th class originally had problems with negative S variances - set S@0 for this model, this then resulted in non-convergence.
Significance values for 2 class model: Q has significant mean for class 1 (0.044), although only just. Q has non significant mean for class 2 (0.085).
Significance values for 3 class model: Q has significant mean for class 1 (0.001). Q has significant mean for class 2 (0.002). Q has significant mean for class 3 (0.008).
# two class
quad.mean.two.class <- si.quad.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.quadratic..isolation_2traj_full_sample_clustered_QUAD.out$parameters$unstandardized %>%
filter(paramHeader == "Means" & LatentClass < 7) # filter out the means for I, S and Q for each class
quad.variance.two.class <- si.quad.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.quadratic..isolation_2traj_full_sample_clustered_QUAD.out$parameters$unstandardized %>%
filter(paramHeader == "Variances" & LatentClass < 7) # filter out the means for I, S and Q for each class
# three class
quad.mean.three.class <- si.quad.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.quadratic..isolation_3traj_full_sample_clustered_QUAD.out$parameters$unstandardized %>%
filter(paramHeader == "Means" & LatentClass < 7) # filter out the means for I, S and Q for each class
quad.variance.three.class <- si.quad.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.quadratic..isolation_3traj_full_sample_clustered_QUAD.out$parameters$unstandardized %>%
filter(paramHeader == "Variances" & LatentClass < 7) # filter out the means for I, S and Q for each class Two class model
two.class.mean.var <- rbind(quad.mean.two.class, quad.variance.two.class) %>%
select(`Mean/variance` = paramHeader,
`Intercept/slope/quadratic` = param,
`estimate` = est,
`standard error` = se,
`p value` = pval)
kable(two.class.mean.var)| Mean/variance | Intercept/slope/quadratic | estimate | standard error | p value |
|---|---|---|---|---|
| Means | I | 0.714 | 0.030 | 0.000 |
| Means | S | 0.033 | 0.022 | 0.127 |
| Means | Q | -0.006 | 0.003 | 0.044 |
| Means | I | 1.979 | 0.227 | 0.000 |
| Means | S | -0.087 | 0.254 | 0.732 |
| Means | Q | 0.067 | 0.039 | 0.085 |
| Variances | I | 0.642 | 0.119 | 0.000 |
| Variances | S | 0.081 | 0.029 | 0.006 |
| Variances | Q | 0.001 | 0.001 | 0.013 |
| Variances | I | 0.642 | 0.119 | 0.000 |
| Variances | S | 0.081 | 0.029 | 0.006 |
| Variances | Q | 0.001 | 0.001 | 0.013 |
Three class model
three.class.mean.var <- rbind(quad.mean.three.class, quad.variance.three.class) %>%
select(`Mean/variance` = paramHeader,
`Intercept/slope/quadratic` = param,
`estimate` = est,
`standard error` = se,
`p value` = pval)
kable(three.class.mean.var)| Mean/variance | Intercept/slope/quadratic | estimate | standard error | p value |
|---|---|---|---|---|
| Means | I | 0.594 | 0.041 | 0.000 |
| Means | S | 0.078 | 0.023 | 0.001 |
| Means | Q | -0.010 | 0.003 | 0.001 |
| Means | I | 1.417 | 0.223 | 0.000 |
| Means | S | -0.142 | 0.207 | 0.493 |
| Means | Q | 0.095 | 0.031 | 0.002 |
| Means | I | 4.274 | 0.638 | 0.000 |
| Means | S | -0.826 | 0.202 | 0.000 |
| Means | Q | 0.069 | 0.026 | 0.008 |
| Variances | I | 0.244 | 0.095 | 0.010 |
| Variances | S | 0.073 | 0.028 | 0.010 |
| Variances | Q | 0.001 | 0.000 | 0.014 |
| Variances | I | 0.244 | 0.095 | 0.010 |
| Variances | S | 0.073 | 0.028 | 0.010 |
| Variances | Q | 0.001 | 0.000 | 0.014 |
| Variances | I | 0.244 | 0.095 | 0.010 |
| Variances | S | 0.073 | 0.028 | 0.010 |
| Variances | Q | 0.001 | 0.000 | 0.014 |
Model fit statistics for two and three class models
# remove 4,5,6 class models
si.two_three.summaries <- si_quad_summaries[-c(3:5),] #social isolation summaries
si.class.summaries <- data.frame(matrix(nrow = 2,
ncol = 8))
#create column names for the variables you will be adding to the empty matrix of si.class.summaries
colnames(si.class.summaries) <- c("Model",
"Parameters",
"AIC",
"BIC",
"aBIC",
"Entropy",
"VLMR LRT p-value",
"Class Proportions") #or whichever indices you want to compare; do si_summaries$ to see what's in there
#check colnames
#si.class.summaries
#create "Model" variable
si.class.summaries <- si.class.summaries %>%
mutate(
Model =
as.factor(c("Two Class", #change nrow in data frame above when adding more models
"Three Class")))
#add summary information into data frame
si.class.summaries <- si.class.summaries %>%
mutate(
Parameters =
si.two_three.summaries$Parameters) %>% #parameters col from original summaries data set
mutate(
AIC =
si.two_three.summaries$AIC) %>% #AIC col from original summaries data set
mutate(
BIC =
si.two_three.summaries$BIC) %>% #BIC col from original summaries data set
mutate(
aBIC =
si.two_three.summaries$aBIC) %>% #aBIC col from original summaries data set
mutate(
Entropy =
si.two_three.summaries$Entropy) %>% #Entropy col from original summaries data set
mutate(
`VLMR LRT p-value` =
si.two_three.summaries$T11_VLMR_PValue) #T11_VLMR_PValue col from original summaries data set
#check
#si.class.summaries#for each class we want to add in the model estimated class counts - then convert this into a list with the percentage of people in each class
#two classes
si.class.summaries$`Class Proportions`[si.class.summaries$Model == "Two Class"] <- list(c(sprintf("%.0f", si.quad.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.quadratic..isolation_2traj_full_sample_clustered_QUAD.out$class_counts$modelEstimated$proportion*100)))
#three classes
si.class.summaries$`Class Proportions`[si.class.summaries$Model == "Three Class"] <- list(c(sprintf('%.0f', si.quad.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.quadratic..isolation_3traj_full_sample_clustered_QUAD.out$class_counts$modelEstimated$proportion*100)))#two classes
si.class.summaries$`Class N`[si.class.summaries$Model == "Two Class"] <- list(c(sprintf("%.0f", si.quad.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.quadratic..isolation_2traj_full_sample_clustered_QUAD.out$class_counts$posteriorProb$count)))
#three classes
si.class.summaries$`Class N`[si.class.summaries$Model == "Three Class"] <- list(c(sprintf("%.0f", si.quad.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.quadratic..isolation_3traj_full_sample_clustered_QUAD.out$class_counts$posteriorProb$count)))#Look at summary table
kable(si.class.summaries)| Model | Parameters | AIC | BIC | aBIC | Entropy | VLMR LRT p-value | Class Proportions | Class N |
|---|---|---|---|---|---|---|---|---|
| Two Class | 17 | 25392.55 | 25489.63 | 25435.62 | 0.950 | 0.0562 | 93, 7 | 2071, 161 |
| Three Class | 21 | 24762.68 | 24882.60 | 24815.88 | 0.956 | 0.2802 | 90, 5 , 5 | 2014, 114 , 104 |
Plots
Two Class model
#create two class data frame with means and variances
two_class_si <- as.data.frame(
si.quad.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.quadratic..isolation_2traj_full_sample_clustered_QUAD.out$gh5$means_and_variances_data$y_estimated_means)
#name the variables
names(two_class_si) <- c("Class 1","Class 2")
#create time point column
two_class_si$timepoint <- c(5, 7, 10, 12)
#convert data to long format
two_class_si <- two_class_si %>%
pivot_longer(-timepoint,
names_to = "Class",
values_to = "social.isolation_score")
#check
#two_class_sitwo_traj_plot_clustered_full_sample <- ggplot(two_class_si,
aes(x = timepoint,
y = social.isolation_score,
colour = Class)) +
geom_line(size = 1.5) +
geom_point(aes(#shape = class,
size = 1)) +
scale_fill_manual(values = palette2) +
scale_color_manual(values = palette2) +
labs(x = "Age (in years)",
title = "Two class model",
color = "Class",
y ="Social isolation score") +
scale_y_continuous(expand = c(0.1,0.1),
limits = c(0,12),
breaks = seq(0, 12, 2)) +
scale_x_continuous(expand = c(0.1,0.1),
limits = c(5,12),
breaks = c(5, 7, 10, 12)) +
theme(panel.grid.major.y = element_line(size = 0.5,
linetype = 'dashed',
colour = "gray"),
axis.text = element_text(colour="black",
size = 12),
axis.title = element_text(colour="black",
size = 12),
panel.background = element_blank(),
plot.title = element_text(size = 16, hjust = 0.5)) +
guides(shape = FALSE, size = FALSE)
two_traj_plot_clustered_full_sample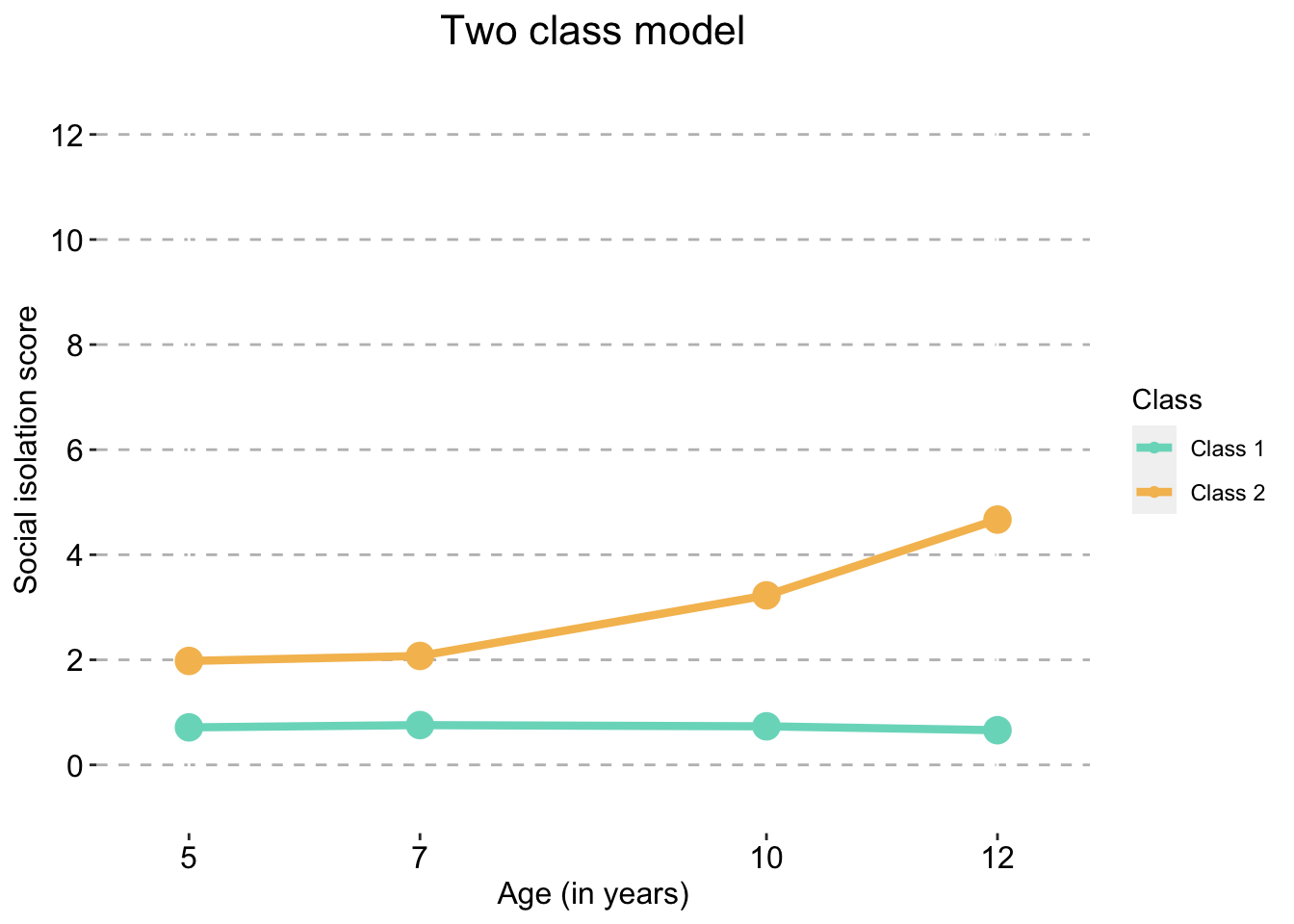
Three Class model
#create three class data frame with means and variances
three_class_si <- as.data.frame(
si.quad.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.quadratic..isolation_3traj_full_sample_clustered_QUAD.out$gh5$means_and_variances_data$y_estimated_means)
#name the variables
names(three_class_si) <- c("Class 1","Class 2","Class 3")
#create time point column
three_class_si$timepoint <- c(5, 7, 10, 12)
#convert data to long format
three_class_si <- three_class_si %>%
pivot_longer(-timepoint,
names_to = "Class",
values_to = "social.isolation_score")
#check
#three_class_sithree_traj_plot_clustered_full_sample <- ggplot(three_class_si,
aes(x = timepoint,
y = social.isolation_score,
colour = Class)) +
geom_line(size = 1.5) +
geom_point(aes(size = 1)) +
scale_fill_manual(values = palette3) +
scale_color_manual(values = palette3) +
labs(x = "Age (in years)",
title = "Three class model",
color = "Class",
y ="Social isolation score") +
scale_y_continuous(expand = c(0.1,0.1),
limits = c(0,12),
breaks = seq(0, 12, 2)) +
scale_x_continuous(expand = c(0.1,0.1),
limits = c(5,12),
breaks = c(5, 7, 10, 12)) +
theme(panel.grid.major.y = element_line(size = 0.5,
linetype = 'dashed',
colour = "gray"),
axis.text = element_text(colour="black",
size = 12),
axis.title = element_text(colour="black",
size = 12),
panel.background = element_blank(),
plot.title = element_text(size = 16, hjust=0.5)) +
guides(shape = FALSE, size = FALSE)
three_traj_plot_clustered_full_sample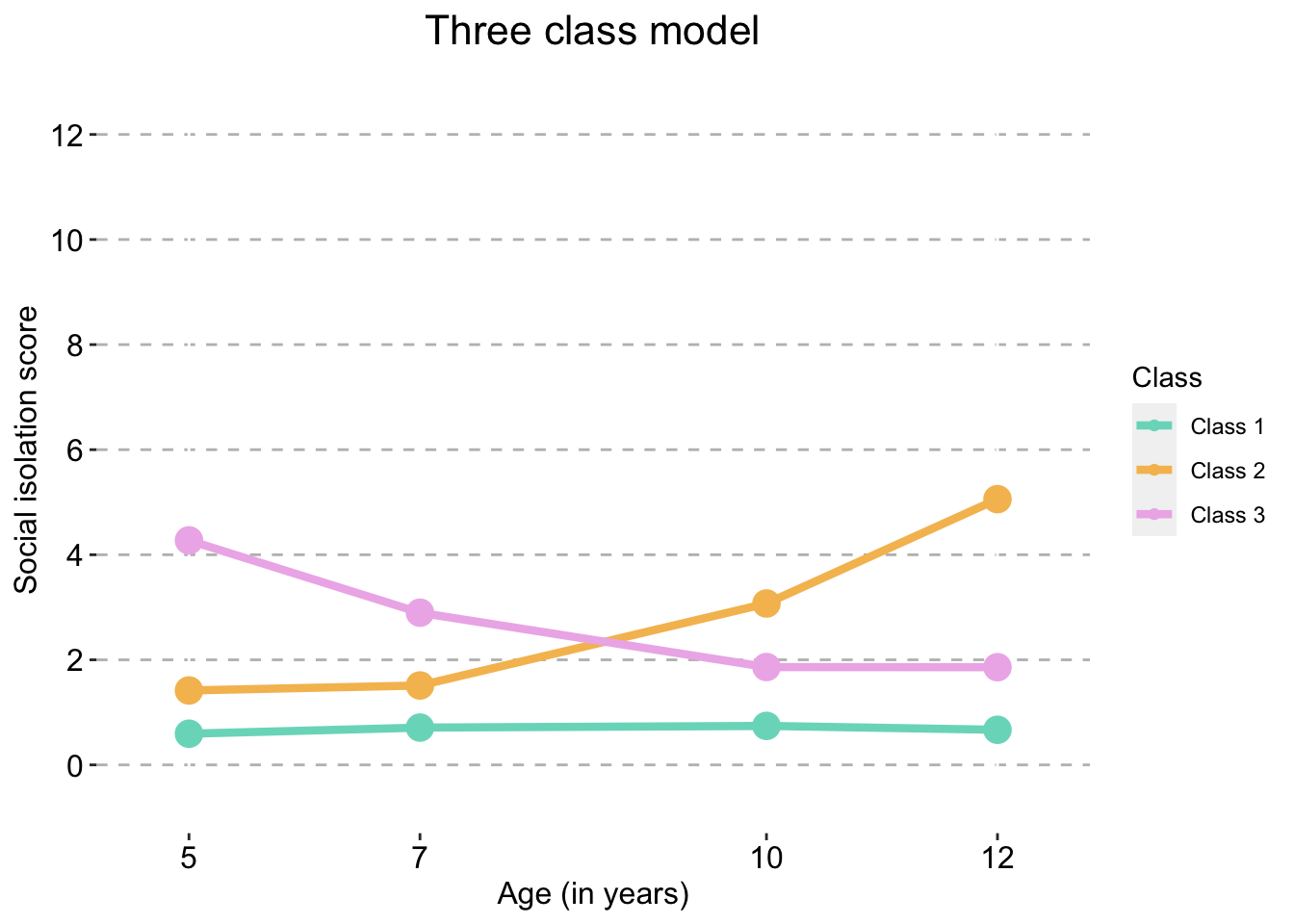
Allocation differences between linear and quadratic 3 class model
dat.descriptives <- readRDS(file = paste0(data_path, "preprocessed_isolation_trajectories_Jan2021_full_sample.rds"))
#colnames(dat.descriptives)dat.tra.prob.full.LINEAR <- read.table(paste0(mplus_GMM_clustered_full_output_data_path, "GMM_SI_3Cl_full_sample_clustered.txt"),
header = FALSE,
col.names = c("SISOE5", # original social isolation score at age 5
"SISOE7", # original social isolation score at age 7
"SISOE10", # original social isolation score at age 10
"SISOE12", # original social isolation score at age 12
"I", # intercept
"S", # slope
"C_I", # factor scores based on most likely class membership
"C_S", # factor scores based on most likely class membership
"CPROB1", # class probability 1
"CPROB2", # class probability 2
"CPROB3", # class probability 3
"C", # class
"ID", # ID
"FAMILYID" # Family ID - twin clustering
))
#check
#dat.tra.prob.full.LINEARdat.tra.prob.full.QUAD <- read.table(paste0(mplus_GMM_clustered_full_output_QUAD_data_path, "GMM_SI_3Cl_full_sample_clustered_QUAD.txt"),
header = FALSE,
col.names = c("SISOE5", # original social isolation score at age 5
"SISOE7", # original social isolation score at age 7
"SISOE10", # original social isolation score at age 10
"SISOE12", # original social isolation score at age 12
"I", # intercept
"S", # slope
"Q", # quadratic
"C_I", # factor scores based on most likely class membership
"C_S", # factor scores based on most likely class membership
"C_Q", # factor scores based on most likely class membership
"CPROB1", # class probability 1
"CPROB2", # class probability 2
"CPROB3", # class probability 3
"C", # class
"ID", # ID
"FAMILYID" # Family ID - twin clustering
))
#check
#dat.tra.prob.full.QUADdat.tra.prob.LINEAR <- dat.tra.prob.full.LINEAR %>%
select(id = ID,
prob1.linear = CPROB1,
prob2.linear = CPROB2,
prob3.linear = CPROB3,
class.linear = C
)
#check
#dat.tra.prob.LINEAR
dat.tra.prob.QUAD <- dat.tra.prob.full.QUAD %>%
select(id = ID,
prob1.quad = CPROB1,
prob2.quad = CPROB2,
prob3.quad = CPROB3,
class.quad = C
)
#check
#dat.tra.prob.QUAD
#create list of data frames to merge
dataframe_list <- list(
dat.descriptives,
dat.tra.prob.LINEAR,
dat.tra.prob.QUAD
)
#merge data frames
dat <- plyr::join_all(
dataframe_list,
by = "id" # Alternatively you can join by several columns
)
#check
#colnames(dat)# linear
dat <- dat %>%
mutate(class_renamed.linear =
recode_factor(class.linear,
"1" = "Low stable",
"2" = "Increasing",
"3" = "Decreasing"
)
)
as.data.frame(table(dat$class_renamed.linear)) %>%
select(`Linear classes` = Var1,
Freq)# quadratic
dat <- dat %>%
mutate(class_renamed.quad =
recode_factor(class.quad,
"1" = "Low stable",
"2" = "Increasing",
"3" = "Decreasing"
)
)
as.data.frame(table(dat$class_renamed.quad)) %>%
select(`Quadratic classes` = Var1,
Freq)Matching classification
96.51% of people were classified the same way regardless if the model was linear or quadratic.
Overall average movement from Linear to Quadratic:
- 1 more person in the increasing class
- 18 less people in the decreasing class
- 17 more people in the low stable class
dat <- dat %>%
mutate(
matching.class =
case_when(
class.linear == class.quad ~ "Matching",
class.linear != class.quad ~ "Not matching"
)
)
freq(dat$matching.class,
cumul = FALSE,
display.type = FALSE,
headings = FALSE,
style = "rmarkdown",
report.nas = FALSE)| Freq | % | |
|---|---|---|
| Matching | 2154 | 96.51 |
| Not matching | 78 | 3.49 |
| Total | 2232 | 100.00 |
- 29 in the decreasing class changed classes in the quadratic model.
- 19 in the increasing class changed classes in the quadratic model.
- 30 in the low stable class changed classes in the quadratic model. == 78 who are not matching.
dat <- dat %>%
mutate(matching.isolated =
case_when(
matching.class == "Matching" & class_renamed.linear == "Increasing" ~ "Matching and increasing",
matching.class == "Matching" & class_renamed.linear == "Decreasing" ~ "Matching and decreasing",
matching.class == "Matching" & class_renamed.linear == "Low stable" ~ "Matching and low stable"
))
freq(dat$matching.isolated,
cumul = FALSE,
display.type = FALSE,
headings = FALSE,
style = "rmarkdown",
report.nas = FALSE)| Freq | % | |
|---|---|---|
| Matching and decreasing | 88 | 4.09 |
| Matching and increasing | 87 | 4.04 |
| Matching and low stable | 1979 | 91.88 |
| Total | 2154 | 100.00 |
dat.non.match <- dat %>%
filter(matching.class == "Not matching")# from linear to quadratic model
dat.non.match <- dat.non.match %>%
mutate(
direction.match =
case_when(
class_renamed.linear == "Increasing" & class_renamed.quad == "Low stable" ~ "Increasing to low stable",
class_renamed.linear == "Decreasing" & class_renamed.quad == "Low stable" ~ "Decreasing to low stable",
class_renamed.linear == "Increasing" & class_renamed.quad == "Decreasing" ~ "Increasing to decreasing",
class_renamed.linear == "Decreasing" & class_renamed.quad == "Increasing" ~ "Decreasing to increasing",
class_renamed.linear == "Low stable" & class_renamed.quad == "Increasing" ~ "Low stable to increasing",
class_renamed.linear == "Low stable" & class_renamed.quad == "Decreasing" ~ "Low stable to decreasing",
)
)
freq(dat.non.match$direction.match,
cumul = FALSE,
display.type = FALSE,
headings = FALSE,
style = "rmarkdown",
report.nas = FALSE)| Freq | % | |
|---|---|---|
| Decreasing to increasing | 1 | 1.28 |
| Decreasing to low stable | 28 | 35.90 |
| Increasing to low stable | 19 | 24.36 |
| Low stable to decreasing | 11 | 14.10 |
| Low stable to increasing | 19 | 24.36 |
| Total | 78 | 100.00 |
- 1 person moved from the decreasing to the increasing class.
- 28 people moved from the decreasing to the low stable class.
- 19 people moved from the increasing to the low stable class.
- 11 people moved from the low stable to the decreasing class.
- 19 people moved from the low stable to the increasing class. == 78 non matching.

Work by Katherine N Thompson
katherine.n.thompson@kcl.ac.uk