GMM trajectory statistics and figures
This page displays the analysis results for latent growth curve models (LGCM) and growth mixture models (GMM) run in Mplus. To do this, we have imported output files from Mplus and formatted the statistics and figures in R, using the MplusAutomation package.
To cite MplusAutomation in publications use:
Hallquist, M. N. & Wiley, J. F. (2018). MplusAutomation: An R Package for Facilitating Large-Scale Latent Variable Analyses in Mplus Structural Equation Modeling, 1-18. doi: 10.1080/10705511.2017.1402334.
A BibTeX entry for LaTeX users is
@Article{, title = {{MplusAutomation}: An {R} Package for Facilitating Large-Scale Latent Variable Analyses in {Mplus}}, author = {Michael N. Hallquist and Joshua F. Wiley}, journal = {Structural Equation Modeling}, year = {2018}, pages = {1–18}, doi = {10.1080/10705511.2017.1402334}, url = {https://www.tandfonline.com/doi/full/10.1080/10705511.2017.1402334}, }
Latent growth curve models (LGCM)
Model fit statistics
First read in the Mplus models, remember to check which data set you are reading in.
si.growth.all <- readModels(paste0(mplus_growth_traj_clustered_full_output_data_path), recursive = TRUE)Extract the summary variables from the mplus output files. Assuming there are multiple files in the directory, model summaries could be retained as a data.frame as we have done here.
si_growth_summaries <- do.call("rbind.fill",
sapply(si.growth.all,
"[",
"summaries"))Cut offs and best fitting models are:
- lowest aBIC
- TLI > 0.95
- CFI > 0.95
- RMSEA < 0.05 indicates close fit, and < 0.08 indicates reasonable fit
We chose the Linear model as the best fitting and most parsimonious model.
#social isolation summaries
si.growth.summaries <- data.frame(matrix(nrow = 4,ncol = 8))
#create column names for the variables you will be adding to the empty matrix of si.growth.summaries
colnames(si.growth.summaries) <- c("Model",
"Parameters",
"AIC",
"BIC",
"aBIC",
"CFI",
"TLI",
"RMSEA") #or whichever indices you want to compare; do si_summaries$ to see what's in there
#create "Model" variable
si.growth.summaries <- si.growth.summaries %>%
mutate(
Model =
as.factor(c("Linear growth model",
"Log linear growth model",
"Quadratic growth model",
"Log quadratic growth model")))
#check
# si.growth.summaries
#add summary information into data frame
si.growth.summaries <- si.growth.summaries %>%
mutate(
Parameters =
si_growth_summaries$Parameters) %>% #parameters
mutate(
AIC =
si_growth_summaries$AIC) %>% #AIC
mutate(
BIC =
si_growth_summaries$BIC) %>% #BIC
mutate(
aBIC =
si_growth_summaries$aBIC) %>% #aBIC
mutate(
CFI =
si_growth_summaries$CFI) %>% #CFI
mutate(
TLI =
si_growth_summaries$TLI) %>% #TLI
mutate(
RMSEA =
si_growth_summaries$RMSEA_Estimate) #RMSEA
#check
# si.growth.summariesknitr::kable(si.growth.summaries) #table for Word| Model | Parameters | AIC | BIC | aBIC | CFI | TLI | RMSEA |
|---|---|---|---|---|---|---|---|
| Linear growth model | 9 | 26268.18 | 26319.58 | 26290.98 | 0.991 | 0.989 | 0.027 |
| Log linear growth model | 9 | 11069.13 | 11120.53 | 11091.93 | 0.993 | 0.992 | 0.030 |
| Quadratic growth model | 13 | 26255.23 | 26329.47 | 26288.16 | 0.997 | 0.984 | 0.033 |
| Log quadratic growth model | 13 | 11064.84 | 11139.08 | 11097.77 | 0.997 | 0.984 | 0.043 |
Means and variances
# linear
linear.mean <- si.growth.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.growth_trajectories.clustered.full_sample.output..isolation_1traj_linear_full_sample_clustered.out$parameters$unstandardized %>%
filter(paramHeader == "Means") # filter out the means for I and S
linear.variance <- si.growth.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.growth_trajectories.clustered.full_sample.output..isolation_1traj_linear_full_sample_clustered.out$parameters$unstandardized %>%
filter(paramHeader == "Variances") # filter out the variance for I and S
# quadratic
quad.mean <- si.growth.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.growth_trajectories.clustered.full_sample.output..isolation_1traj_quad_full_sample_clustered.out$parameters$unstandardized %>%
filter(paramHeader == "Means") # filter out the means for I, S and Q
quad.variance <- si.growth.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.growth_trajectories.clustered.full_sample.output..isolation_1traj_quad_full_sample_clustered.out$parameters$unstandardized %>%
filter(paramHeader == "Variances") # filter out the variance for I, S and Q Linear LCGM means and variances
linear.mean.var <- rbind(linear.mean, linear.variance) %>%
select(`Mean/variance` = paramHeader,
`Intercept/slope` = param,
`estimate` = est,
`standard error` = se,
`p value` = pval)
kable(linear.mean.var)| Mean/variance | Intercept/slope | estimate | standard error | p value |
|---|---|---|---|---|
| Means | I | 0.808 | 0.026 | 0 |
| Means | S | 0.021 | 0.005 | 0 |
| Variances | I | 0.718 | 0.079 | 0 |
| Variances | S | 0.020 | 0.003 | 0 |
Quadratic LCGM means and variances
quad.mean.var <- rbind(quad.mean, quad.variance) %>%
select(`Mean/variance` = paramHeader,
`Intercept/slope/quadratic` = param,
`estimate` = est,
`standard error` = se,
`p value` = pval)
kable(quad.mean.var)| Mean/variance | Intercept/slope/quadratic | estimate | standard error | p value |
|---|---|---|---|---|
| Means | I | 0.806 | 0.027 | 0.000 |
| Means | S | 0.025 | 0.015 | 0.093 |
| Means | Q | -0.001 | 0.002 | 0.773 |
| Variances | I | 0.788 | 0.127 | 0.000 |
| Variances | S | 0.090 | 0.029 | 0.002 |
| Variances | Q | 0.001 | 0.000 | 0.002 |
Plots
Linear and quadratic growth curves
#create empty dataset
dat_si <- data.frame(matrix(nrow = 4, ncol = 3))
#add column names
colnames(dat_si) <- c("Observed", "Linear", "Quadratic")
#add in model statistics from out files - will need to change the folder names depending on which sample you are looking at
dat_si$Observed <- si.growth.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.growth_trajectories.clustered.full_sample.output..isolation_1traj_linear_full_sample_clustered.out$gh5$means_and_variances_data$y_observed_means$values #observed means - used linear but could be any
dat_si$Linear <- si.growth.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.growth_trajectories.clustered.full_sample.output..isolation_1traj_linear_full_sample_clustered.out$gh5$means_and_variances_data$y_estimated_means$values #estimated means
dat_si$Quadratic <- si.growth.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.growth_trajectories.clustered.full_sample.output..isolation_1traj_quad_full_sample_clustered.out$gh5$means_and_variances_data$y_estimated_means$values #estimated means
#add timepoint column
dat_si$Timepoint <- c(5, 7, 10, 12)
#convert into long format
dat_si <- dat_si %>%
tidyr::pivot_longer(-Timepoint,
names_to = "Model",
values_to = "score")
#check
dat_siisolation_growth_curve_clustered_full_sample <- ggplot() + #change name of graph here if using the 3 missing sample
geom_line(data = dat_si,
aes(x = Timepoint,
y = score,
colour = Model),
size = 1) +
geom_point(data = dat_si,
aes(x = Timepoint,
y = score,
colour = Model,
shape = Model),
show.legend = T) +
scale_color_manual(values = palette) +
labs(x = "Age (years)",
y ="Social isolation",
title = "Growth curves for social isolation") +
scale_y_continuous(expand = c(0.01,0.01),
limits = c(0.8,0.96),
breaks=seq(0.8, 0.96, 0.02)) +
scale.x +
combined.traj.theme
isolation_growth_curve_clustered_full_sample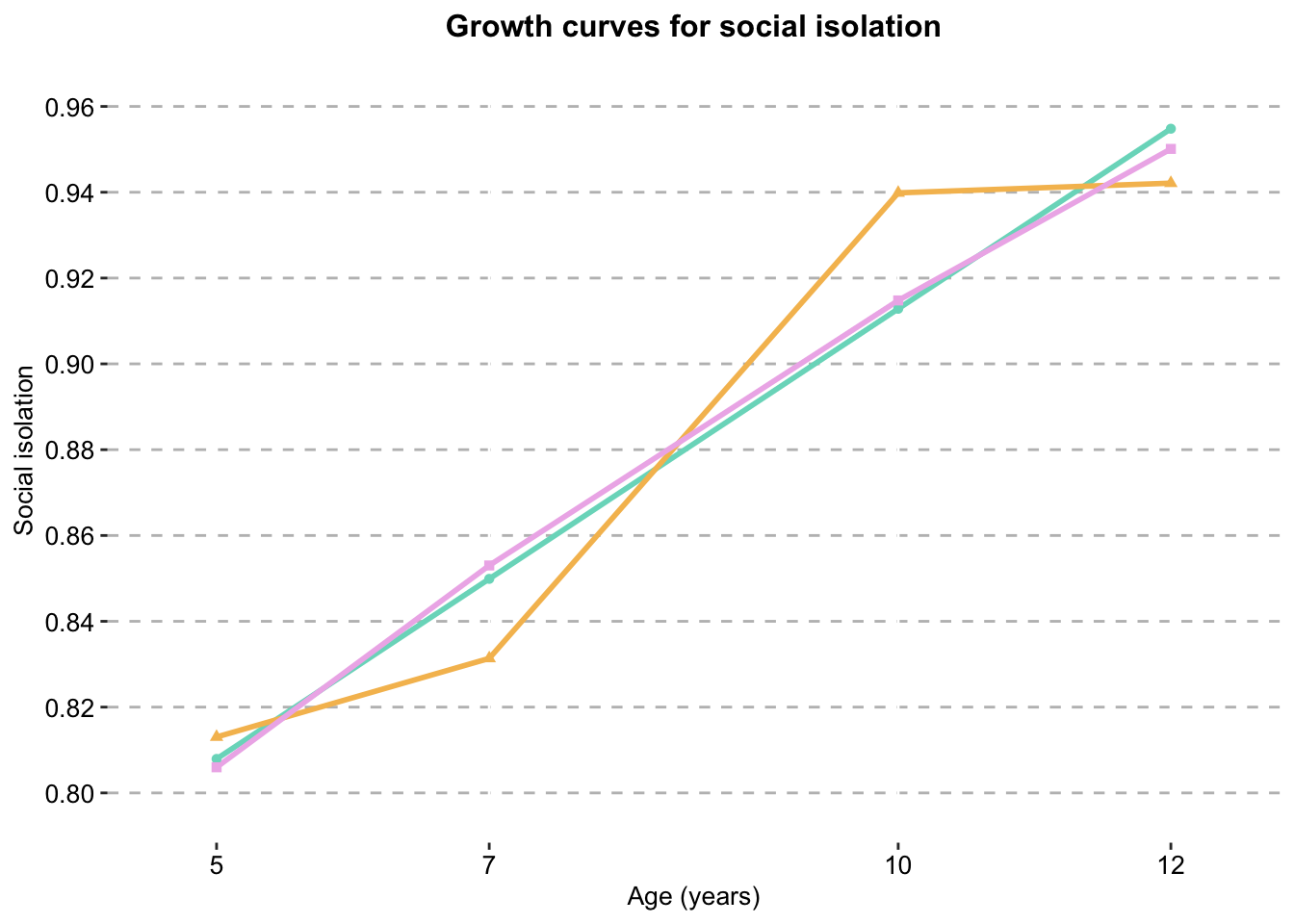
Linear growth curve only
#create empty dataset
dat_si_single <- data.frame(matrix(nrow = 4, ncol = 1))
#add column names
colnames(dat_si_single) <- c("Linear growth curve")
#add in model statistics from out files - will need to change the folder names depending on which sample
dat_si_single$`Linear growth curve` <- si.growth.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.growth_trajectories.clustered.full_sample.output..isolation_1traj_linear_full_sample_clustered.out$gh5$means_and_variances_data$y_estimated_means$values #estimated means
#add timepoint column
dat_si_single$timepoint <- c(5, 7, 10, 12)
#convert into long format
dat_si_single <- dat_si_single %>%
tidyr::pivot_longer(-timepoint,
names_to = "Class",
values_to = "social.isolation_score")
#check
#dat_si_singleisolation_growth_curve_single_clustered_full_sample <- ggplot() +
geom_line(data = dat_si_single,
aes(x = timepoint,
y = social.isolation_score,
# colour = Class,
shape = Class),
size = 1.5) +
geom_point(data = dat_si_single,
aes(x = timepoint,
y = social.isolation_score,
# colour = Class,
size = 1),
show.legend = T) +
scale_color_manual(values = palette) +
labs(x = "Age (years)",
y ="Social isolation",
title = "Linear latent growth curve",
color = "") +
scale.y +
scale.x +
combined.traj.theme +
guides(shape = FALSE, size = FALSE)
isolation_growth_curve_single_clustered_full_sample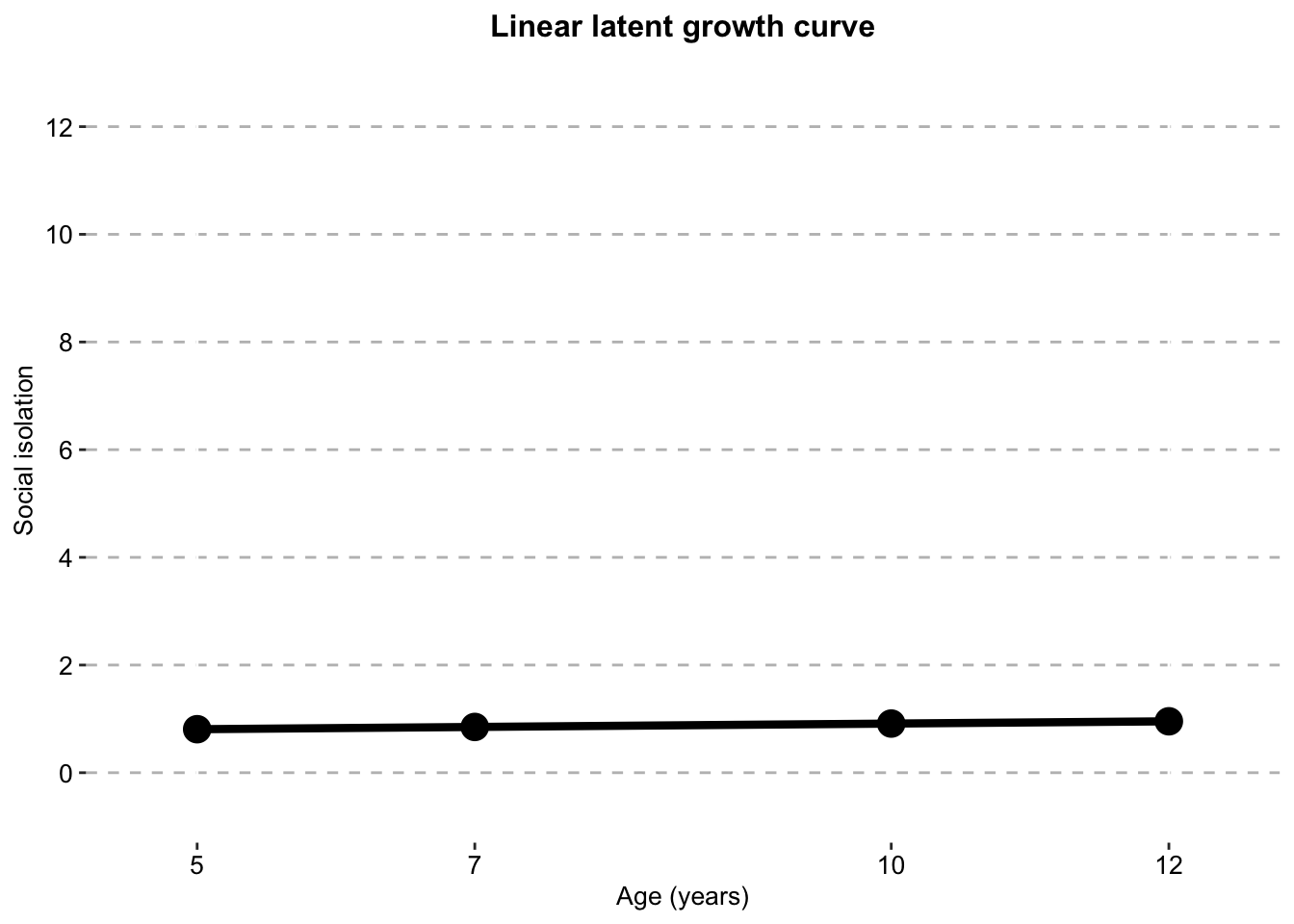
Log-transformed linear and quadratic growth curves
#create empty dataset
dat_si_log <- data.frame(matrix(nrow = 4, ncol = 3))
#add column names
colnames(dat_si_log) <- c("Observed", "Linear", "Quadratic")
#add in model statistics from out files - need to change file name dependant on which sample
dat_si_log$Observed <- si.growth.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.growth_trajectories.clustered.full_sample.output..isolation_1traj_linear_log_full_sample_clustered.out$gh5$means_and_variances_data$y_observed_means$values #observed means
dat_si_log$Linear <- si.growth.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.growth_trajectories.clustered.full_sample.output..isolation_1traj_linear_log_full_sample_clustered.out$gh5$means_and_variances_data$y_estimated_means$values #estimated means
dat_si_log$Quadratic <- si.growth.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.growth_trajectories.clustered.full_sample.output..isolation_1traj_quad_log_full_sample_clustered.out$gh5$means_and_variances_data$y_estimated_means$values #estimated means
#add timepoint column
dat_si_log$Timepoint <- c(5, 7, 10, 12)
#convert into long format
dat_si_log <- dat_si_log %>%
pivot_longer(-Timepoint,
names_to = "Model",
values_to = "score")
#check
#dat_si_logisolation_growth_curve_log_scale_clustered_full_sample <- ggplot() +
geom_line(data = dat_si_log,
aes(x = Timepoint,
y = score,
colour = Model),
size = 1) +
geom_point(data = dat_si_log,
aes(x = Timepoint,
y = score,
colour = Model,
shape=Model),
show.legend = T) +
scale_color_manual(values = palette) +
labs(x = "Age (years)",
y ="Social isolation",
title = "Growth curves for log-social isolation") +
scale_y_continuous(expand = c(0.01,0.01),
limits = c(0.44,0.52),
breaks=seq(0.44, 0.52, 0.01)) +
scale_x_continuous(expand = c(0.1,0.1),
limits = c(5,12),
breaks= c(5, 7, 10, 12)) +
combined.traj.theme
isolation_growth_curve_log_scale_clustered_full_sample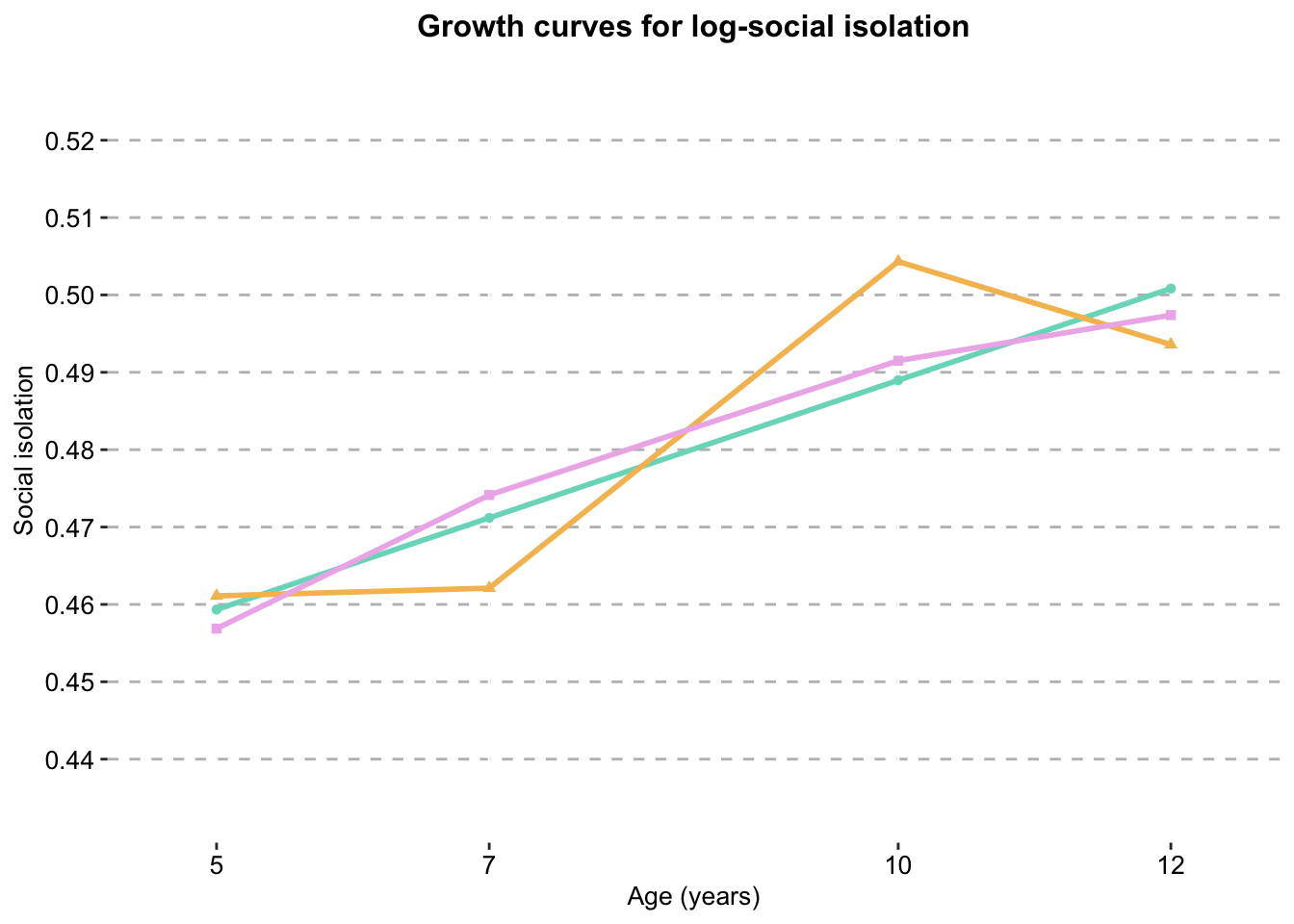
Growth mixture models (GMM)
As the linear curve was the best fit when running a single latent growth curve, all GMM models are lienar form.
Model fit statistics
#For class models, read in all output files within your folder that you have all your class models
si.classes.all <- readModels(paste0(mplus_GMM_clustered_full_output_data_path), recursive = TRUE) #Recursive means it reads data in sub-folders too#extract the summary variables from the mplus output files. Assuming there are multiple files in the above directory, model summaries could be retained as a data.frame as follows:
si_summaries <- do.call("rbind.fill",
sapply(si.classes.all,
"[",
"summaries"))
# have a look at the summary data for all files
si_summaries <- si_summaries[-c(2,3,4,5,7,8,11,13),] # remove rows that are output from OPTSEED tests and keep 5 and 6 trajectory models that fix S to zero#social isolation summaries
si.class.summaries <- data.frame(matrix(nrow = 5,
ncol = 8))
#create column names for the variables you will be adding to the empty matrix of si.class.summaries
colnames(si.class.summaries) <- c("Model",
"Parameters",
"AIC",
"BIC",
"aBIC",
"Entropy",
"VLMR LRT p-value",
"Class Proportions") #or whichever indices you want to compare; do si_summaries$ to see what's in there
#check colnames
#si.class.summaries
#create "Model" variable
si.class.summaries <- si.class.summaries %>%
mutate(
Model =
as.factor(c("Two Class", #change nrow in data frame above when adding more models
"Three Class",
"Four Class",
"Five Class",
"Six Class")))
#add summary information into data frame
si.class.summaries <- si.class.summaries %>%
mutate(
Parameters =
si_summaries$Parameters) %>% #parameters col from original summaries data set
mutate(
AIC =
si_summaries$AIC) %>% #AIC col from original summaries data set
mutate(
BIC =
si_summaries$BIC) %>% #BIC col from original summaries data set
mutate(
aBIC =
si_summaries$aBIC) %>% #aBIC col from original summaries data set
mutate(
Entropy =
si_summaries$Entropy) %>% #Entropy col from original summaries data set
mutate(
`VLMR LRT p-value` =
si_summaries$T11_VLMR_PValue) #T11_VLMR_PValue col from original summaries data set
#check
#si.class.summaries#for each class we want to add in the model estimated class counts - then convert this into a list with the percentage of people in each class
#two classes
si.class.summaries$`Class Proportions`[si.class.summaries$Model == "Two Class"] <- list(c(sprintf("%.0f", si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_2traj_full_sample_clustered.out$class_counts$modelEstimated$proportion*100)))
#three classes
si.class.summaries$`Class Proportions`[si.class.summaries$Model == "Three Class"] <- list(c(sprintf('%.0f', si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_3traj_full_sample_clustered.out$class_counts$modelEstimated$proportion*100)))
#four classes
si.class.summaries$`Class Proportions`[si.class.summaries$Model == "Four Class"] <- list(c(sprintf('%.0f', si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_4traj_full_sample_clustered.out$class_counts$modelEstimated$proportion*100)))
#five classes
si.class.summaries$`Class Proportions`[si.class.summaries$Model == "Five Class"] <- list(c(sprintf('%.0f', si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_5traj_full_sample_clustered_S_fixed_to_zero.out$class_counts$modelEstimated$proportion*100)))
#six classes
si.class.summaries$`Class Proportions`[si.class.summaries$Model == "Six Class"] <- list(c(sprintf('%.0f', si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_6traj_full_sample_clustered_S_fixed_to_zero.out$class_counts$modelEstimated$proportion*100)))#two classes
si.class.summaries$`Class N`[si.class.summaries$Model == "Two Class"] <- list(c(sprintf("%.0f", si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_2traj_full_sample_clustered.out$class_counts$posteriorProb$count)))
#three classes
si.class.summaries$`Class N`[si.class.summaries$Model == "Three Class"] <- list(c(sprintf("%.0f", si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_3traj_full_sample_clustered.out$class_counts$posteriorProb$count)))
#four classes
si.class.summaries$`Class N`[si.class.summaries$Model == "Four Class"] <- list(c(sprintf("%.0f", si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_4traj_full_sample_clustered.out$class_counts$posteriorProb$count)))
#five classes
si.class.summaries$`Class N`[si.class.summaries$Model == "Five Class"] <- list(c(sprintf("%.0f", si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_5traj_full_sample_clustered_S_fixed_to_zero.out$class_counts$posteriorProb$count)))
#six classes
si.class.summaries$`Class N`[si.class.summaries$Model == "Six Class"] <- list(c(sprintf("%.0f", si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_6traj_full_sample_clustered_S_fixed_to_zero.out$class_counts$posteriorProb$count)))The three class model is chosen as the best fitting model according to fit statistics and logical evaluation The five and six class models here have fixed the variance of S (the slope) to zero, as the original model produces negative variances and is not interpretable.
#Look at summary table
kable(si.class.summaries)| Model | Parameters | AIC | BIC | aBIC | Entropy | VLMR LRT p-value | Class Proportions | Class N |
|---|---|---|---|---|---|---|---|---|
| Two Class | 12 | 25447.50 | 25516.03 | 25477.90 | 0.951 | 0.0344 | 93, 7 | 2077, 155 |
| Three Class | 15 | 24922.10 | 25007.76 | 24960.11 | 0.952 | 0.0587 | 90, 5 , 5 | 2007, 110 , 115 |
| Four Class | 18 | 24629.97 | 24732.76 | 24675.57 | 0.936 | 0.8906 | 4 , 84, 2 , 11 | 83 , 1874, 36 , 238 |
| Five Class | 19 | 24352.62 | 24461.12 | 24400.76 | 0.942 | 0.0541 | 5 , 82, 1 , 1 , 10 | 110 , 1838, 33 , 20 , 232 |
| Six Class | 22 | 24163.34 | 24288.98 | 24219.08 | 0.928 | 0.2071 | 1 , 1 , 14, 77, 3 , 4 | 31 , 12 , 309 , 1719, 67 , 94 |
Means and variances
gmm.linear.means.1 <- si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_3traj_full_sample_clustered.out$parameters$unstandardized %>%
filter(paramHeader == "Means" & LatentClass == 1) # filter out the means for I and S
gmm.linear.variances.1 <- si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_3traj_full_sample_clustered.out$parameters$unstandardized %>%
filter(paramHeader == "Variances" & LatentClass == 1) # filter out the variance for I and Sgmm.linear.means.2 <- si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_3traj_full_sample_clustered.out$parameters$unstandardized %>%
filter(paramHeader == "Means" & LatentClass == 2) # filter out the means for I and S
gmm.linear.variances.2 <- si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_3traj_full_sample_clustered.out$parameters$unstandardized %>%
filter(paramHeader == "Variances" & LatentClass == 2) # filter out the variance for I and Sgmm.linear.means.3 <- si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_3traj_full_sample_clustered.out$parameters$unstandardized %>%
filter(paramHeader == "Means" & LatentClass == 3) # filter out the means for I and S
gmm.linear.variances.3 <- si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_3traj_full_sample_clustered.out$parameters$unstandardized %>%
filter(paramHeader == "Variances" & LatentClass == 3) # filter out the variance for I and Sgmm.linear.means.variances <- rbind(gmm.linear.means.1, gmm.linear.variances.1, gmm.linear.means.2, gmm.linear.variances.2, gmm.linear.means.3, gmm.linear.variances.3)%>%
select(Class = LatentClass,
`Means/variances` = paramHeader,
`Intercept/slope` = param,
estimate = est,
`standard error` = se,
`p value` = pval)
kable(gmm.linear.means.variances)| Class | Means/variances | Intercept/slope | estimate | standard error | p value |
|---|---|---|---|---|---|
| 1 | Means | I | 0.612 | 0.031 | 0.000 |
| 1 | Means | S | 0.012 | 0.006 | 0.045 |
| 1 | Variances | I | 0.171 | 0.055 | 0.002 |
| 1 | Variances | S | 0.003 | 0.002 | 0.125 |
| 2 | Means | I | 1.077 | 0.202 | 0.000 |
| 2 | Means | S | 0.541 | 0.062 | 0.000 |
| 2 | Variances | I | 0.171 | 0.055 | 0.002 |
| 2 | Variances | S | 0.003 | 0.002 | 0.125 |
| 3 | Means | I | 3.983 | 0.369 | 0.000 |
| 3 | Means | S | -0.318 | 0.053 | 0.000 |
| 3 | Variances | I | 0.171 | 0.055 | 0.002 |
| 3 | Variances | S | 0.003 | 0.002 | 0.125 |
Plots
Two Class model
#create two class data frame with means and variances
two_class_si <- as.data.frame(
si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_2traj_full_sample_clustered.out$gh5$means_and_variances_data$y_estimated_means)
#name the variables
names(two_class_si) <- c("Class 1","Class 2")
#create timepoint column
two_class_si$timepoint <- c(5, 7, 10, 12)
#convert data to long format
two_class_si <- two_class_si %>%
pivot_longer(-timepoint,
names_to = "Class",
values_to = "social.isolation_score")
# create factor for Class variable
two_class_si <- two_class_si %>%
mutate(Class =
factor(Class,
levels = c("Class 1","Class 2")))
two_class_sitwo_traj_plot_clustered_full_sample <- ggplot(two_class_si,
aes(x = timepoint,
y = social.isolation_score,
# colour = Class,
shape = Class)) +
geom_line(size = 1.5) +
geom_point(aes(size = 1)) +
scale_fill_manual(values = palette) +
scale_color_manual(values = palette) +
labs(x = "Age (years)",
title = "Two trajectory model",
color = "",
y = "Social isolation") +
scale.y +
scale.x +
combined.traj.theme +
guides(shape = FALSE, size = FALSE)
two_traj_plot_clustered_full_sample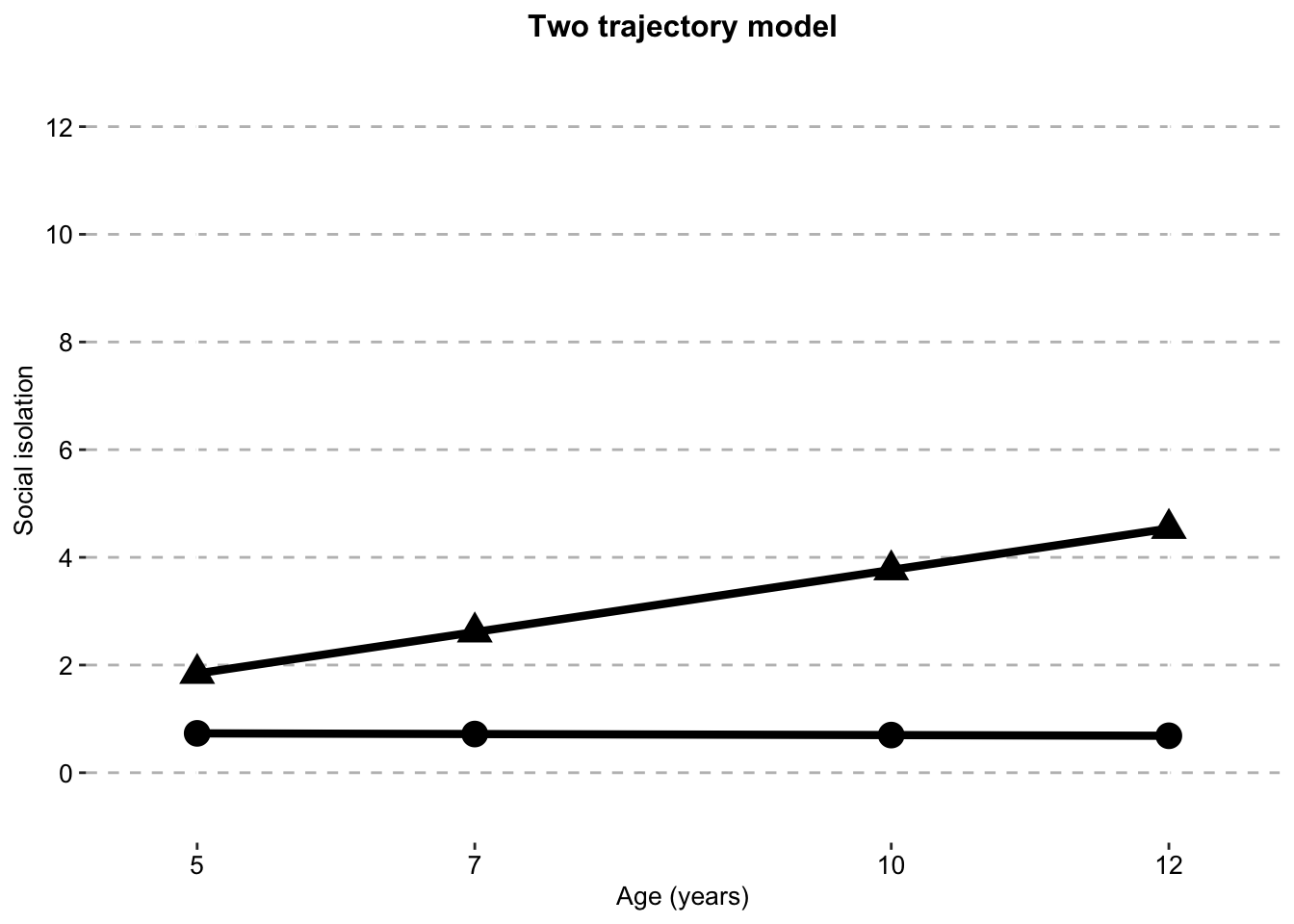
Three Class model
#create three class data frame with means and variances
three_class_si <- as.data.frame(
si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_3traj_full_sample_clustered.out$gh5$means_and_variances_data$y_estimated_means)
#name the variables
names(three_class_si) <- c("Class 1","Class 2","Class 3")
#create time point column
three_class_si$timepoint <- c(5, 7, 10, 12)
#convert data to long format
three_class_si <- three_class_si %>%
pivot_longer(-timepoint,
names_to = "Class",
values_to = "social.isolation_score")
# create factor for Class variable
three_class_si <- three_class_si %>%
mutate(Class =
factor(Class,
levels = c("Class 1","Class 2","Class 3")))
three_class_sithree_traj_plot_clustered_full_sample <- ggplot(three_class_si,
aes(x = timepoint,
y = social.isolation_score,
# colour = Class,
shape = Class)) +
geom_line(size = 1.5) +
geom_point(aes(size = 1)) +
scale_fill_manual(values = palette) +
scale_color_manual(values = palette) +
labs(x = "Age (years)",
title = "Three trajectory model",
color = "",
y ="Social isolation") +
scale.y +
scale.x +
combined.traj.theme +
guides(shape = FALSE, size = FALSE)
three_traj_plot_clustered_full_sample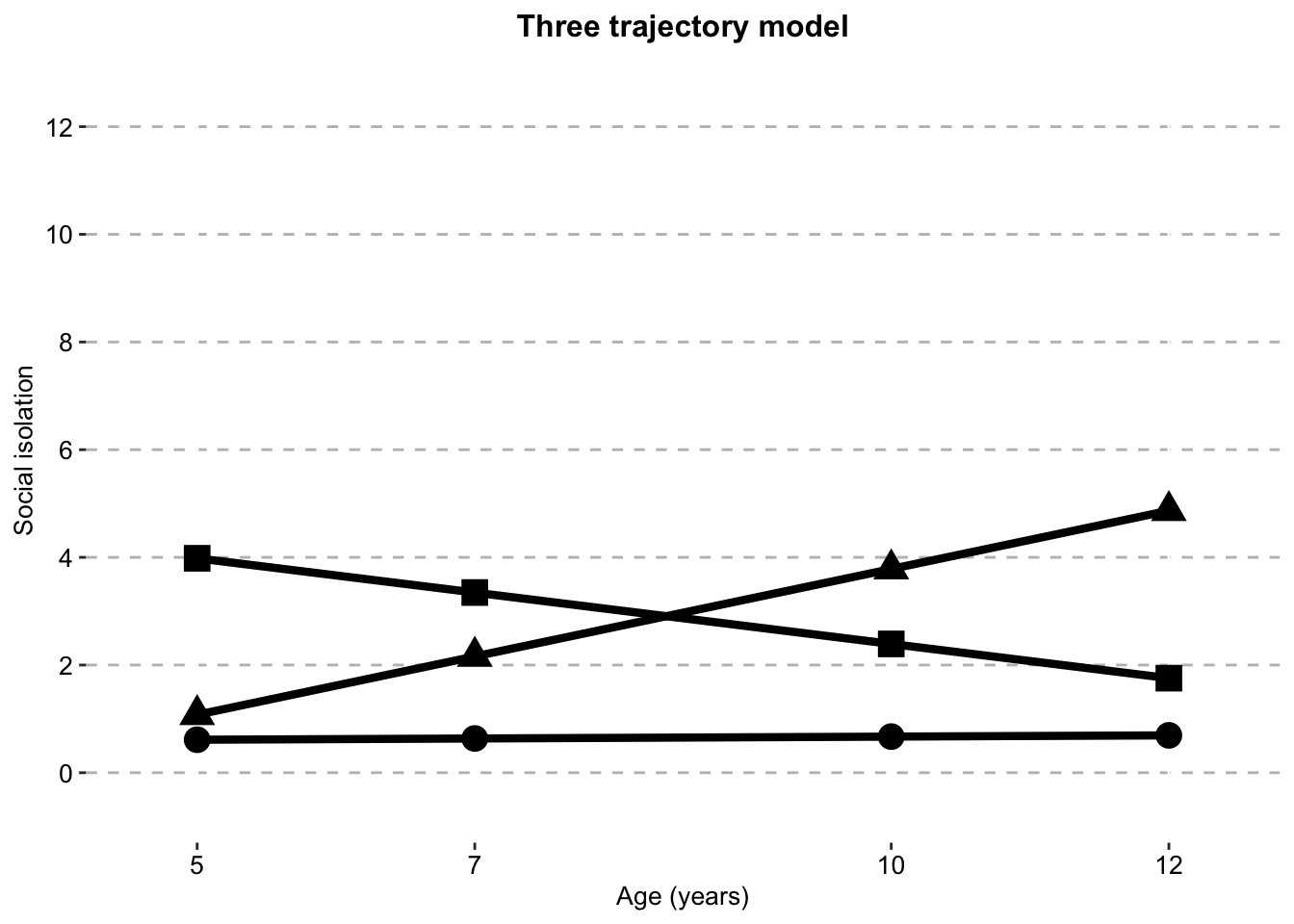
Four Class model
#create four class data frame with means and variances
four_class_si <- as.data.frame(
si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_4traj_full_sample_clustered.out$gh5$means_and_variances_data$y_estimated_means)
#name the variables
names(four_class_si) <- c("Class 1","Class 2","Class 3","Class 4")
#create timepoint column
four_class_si$timepoint <- c(5, 7, 10, 12)
#convert data to long format
four_class_si <- four_class_si %>%
pivot_longer(-timepoint,
names_to = "Class",
values_to = "social.isolation_score")
# create factor for Class variable to order the colour selection in the graph
four_class_si <- four_class_si %>%
mutate(Class =
factor(Class,
levels = c("Class 2",
"Class 4",
"Class 1",
"Class 3")))
four_class_sifour_traj_plot_clustered_full_sample <- ggplot(four_class_si,
aes(x = timepoint,
y = social.isolation_score,
# colour = Class,
shape = Class)) +
geom_line(size = 1.5) +
geom_point(aes(size = 1)) +
scale_fill_manual(values = palette) +
scale_color_manual(values = palette) +
labs(x = "Age (years)",
title = "Four trajectory model",
color = "",
y ="Social isolation") +
scale.y +
scale.x +
combined.traj.theme +
guides(shape = FALSE, size = FALSE)
four_traj_plot_clustered_full_sample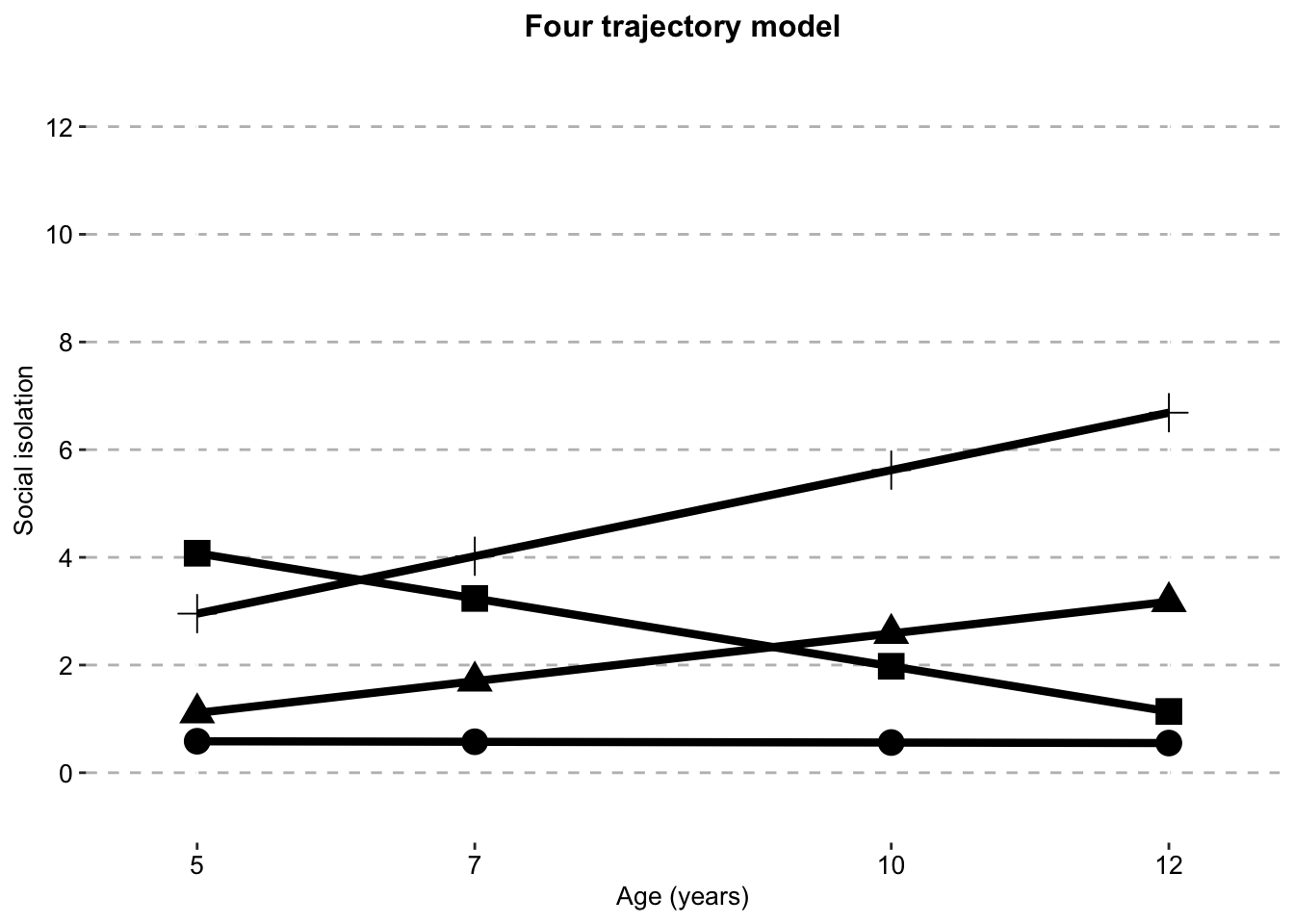
Five Class model
#create five class data frame with means and variances
five_class_si <- as.data.frame(
si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_5traj_full_sample_clustered_S_fixed_to_zero.out$gh5$means_and_variances_data$y_estimated_means)
#name the variables
names(five_class_si) <- c("Class 1","Class 2","Class 3","Class 4","Class 5")
#create timepoint column
five_class_si$timepoint <- c(5, 7, 10, 12)
#convert data to long format
five_class_si <- five_class_si %>%
pivot_longer(-timepoint,
names_to = "Class",
values_to = "social.isolation_score")
# create factor for Class variable
five_class_si <- five_class_si %>%
mutate(Class =
factor(Class,
levels = c("Class 2",
"Class 5",
"Class 1",
"Class 4",
"Class 3")))
five_class_sifive_traj_plot_clustered_full_sample <- ggplot(five_class_si,
aes(x = timepoint,
y = social.isolation_score,
# colour = Class,
shape = Class)) +
geom_line(size = 1.5) +
geom_point(aes(size = 1)) +
scale_fill_manual(values = palette) +
scale_color_manual(values = palette) +
labs(x = "Age (years)",
title = "Five trajectory model",
color = "",
y ="Social isolation") +
scale.y +
scale.x +
combined.traj.theme +
guides(shape = FALSE, size = FALSE)
five_traj_plot_clustered_full_sample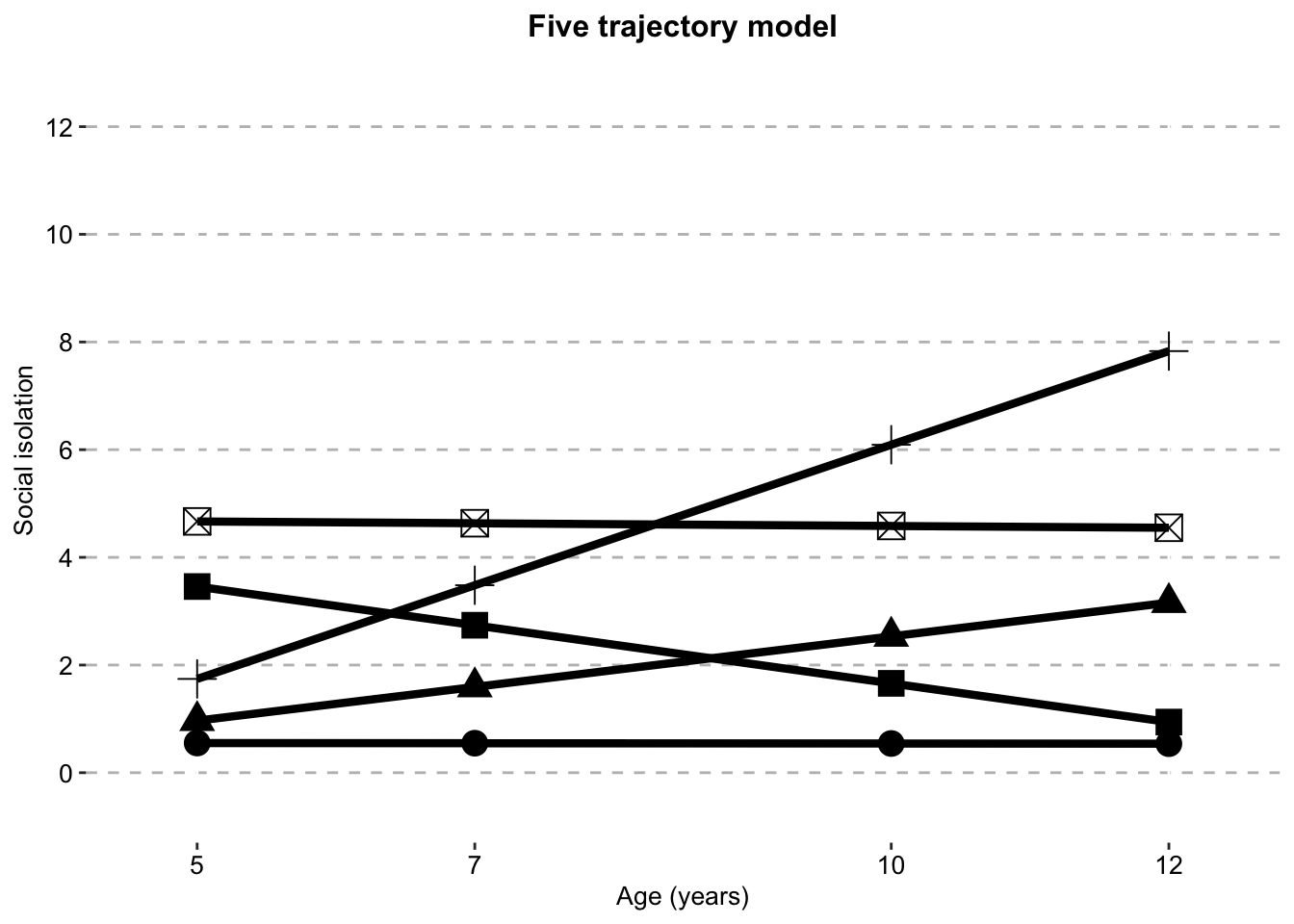
Six Class model
#create five class data frame with means and variances
six_class_si <- as.data.frame(
si.classes.all$X.Users.katiethompson.Documents.PhD.LISS.DTP_Louise_and_Tim.Social.isolation.trajectories_Paper.1.data_analysis.mplus.GMM.clustered.full_sample.output..isolation_6traj_full_sample_clustered_S_fixed_to_zero.out$gh5$means_and_variances_data$y_estimated_means)
#name the variables
names(six_class_si) <- c("Class 1","Class 2","Class 3","Class 4","Class 5","Class 6")
#create timepoint column
six_class_si$timepoint <- c(5, 7, 10, 12)
#convert data to long format
six_class_si <- six_class_si %>%
pivot_longer(-timepoint,
names_to = "Class",
values_to = "social.isolation_score")
six_class_si <- six_class_si %>%
mutate(Class =
factor(Class,
levels = c("Class 4",
"Class 3",
"Class 6",
"Class 2",
"Class 1",
"Class 5")))
six_class_sisix_traj_plot_clustered_full_sample <- ggplot(six_class_si,
aes(x = timepoint,
y = social.isolation_score,
# colour = Class,
shape = Class)) +
geom_line(size = 1.5) +
geom_point(aes(size = 1)) +
scale_fill_manual(values = palette) +
scale_color_manual(values = palette) +
labs(x = "Age (years)",
title = "Six trajectory model",
color = "",
y = "Social isolation") +
scale.y +
scale.x +
combined.traj.theme +
guides(shape = FALSE, size = FALSE)
six_traj_plot_clustered_full_sample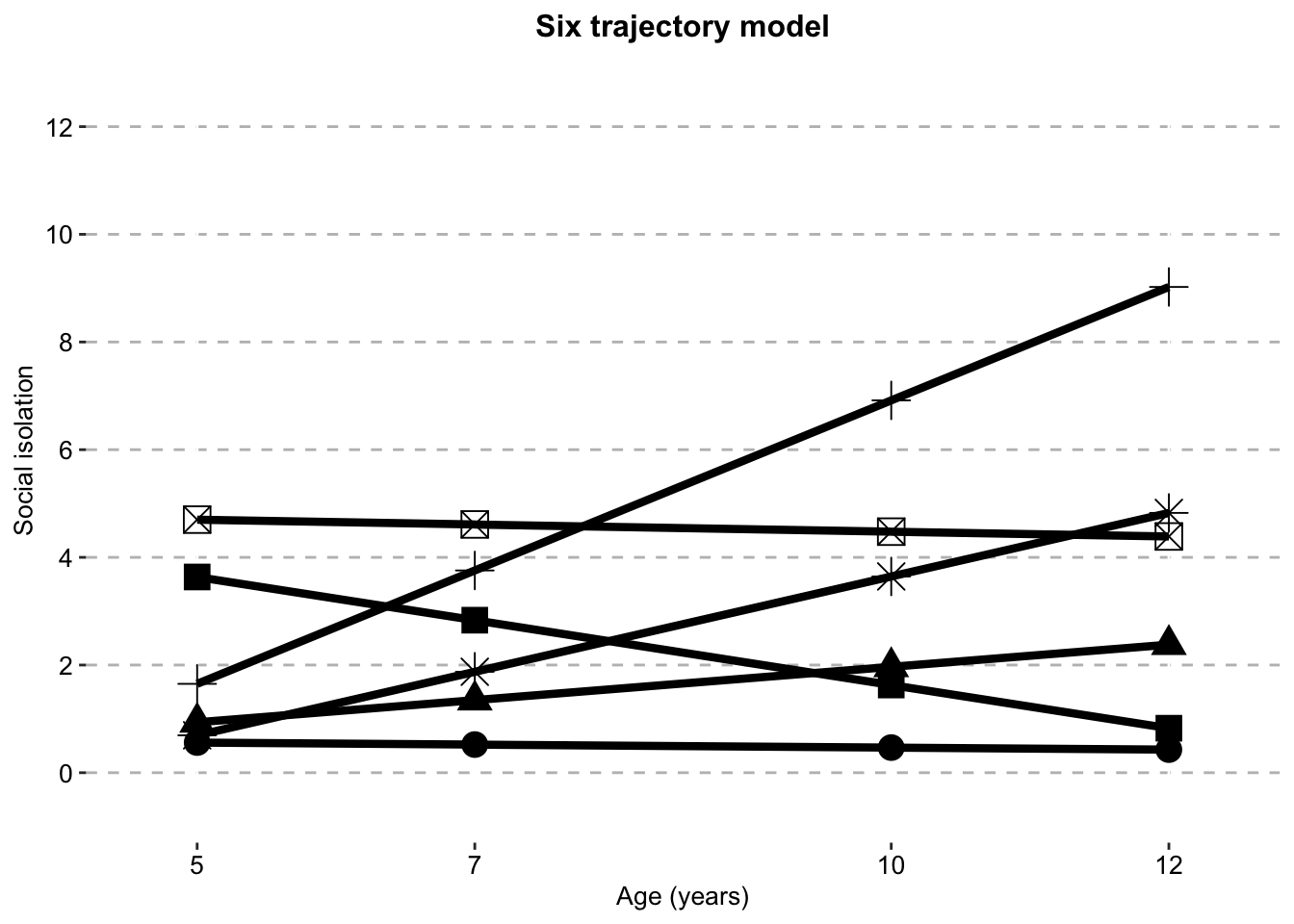
Plot for all models
#combine all plots
library(ggpubr) # Needed for combining plots
library(gridExtra)
combined_trajectories_plot_clustered_full_sample <- ggarrange(isolation_growth_curve_single_clustered_full_sample,
two_traj_plot_clustered_full_sample,
three_traj_plot_clustered_full_sample,
four_traj_plot_clustered_full_sample,
five_traj_plot_clustered_full_sample,
six_traj_plot_clustered_full_sample,
ncol = 2, nrow = 3,
widths = c(2.7,2.7,2.7,2.7),
heights = c(3,3,3,3),
labels = c("A", "B", "C", "D", "E", "F"),
legend = "none"
)
# combined_trajectories_plot_clustered_full_sample_CENTRE_PRES <- ggarrange(isolation_growth_curve_single_clustered_full_sample,
# two_traj_plot_clustered_full_sample,
# three_traj_plot_clustered_full_sample,
# four_traj_plot_clustered_full_sample,
# five_traj_plot_clustered_full_sample,
# six_traj_plot_clustered_full_sample,
# ncol = 3, nrow = 2,
# widths = c(2.7,2.7,2.7,2.7),
# heights = c(3,3,3,3),
# labels = c("A", "B", "C", "D", "E", "F"),
# legend = "none"
# )combined_trajectories_plot_clustered_full_sample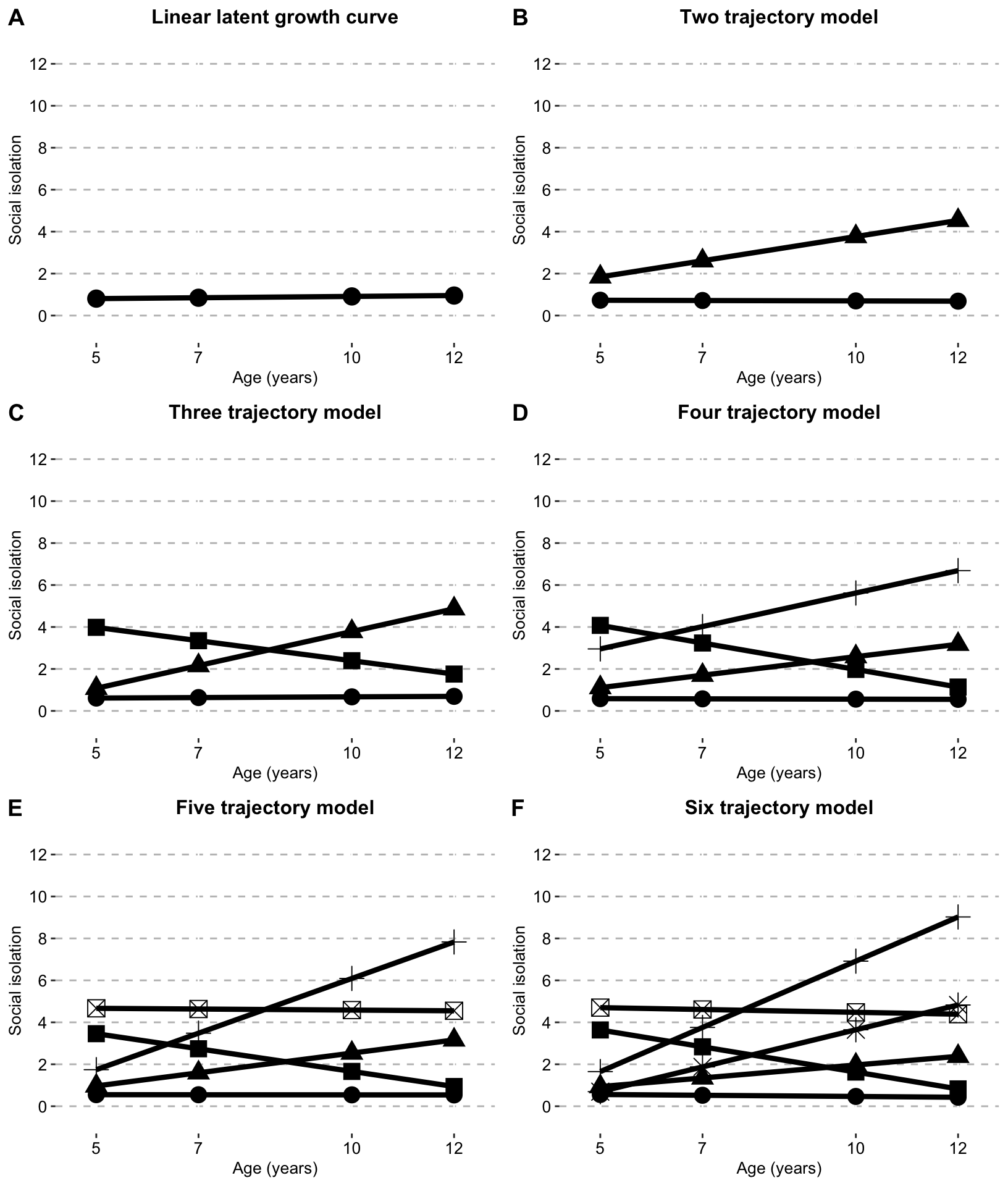

Work by Katherine N Thompson
katherine.n.thompson@kcl.ac.uk